With the help of this utility, you can perform missing data imputation on a PC. You have the option to enter command line arguments and specify parameters.
Beagle
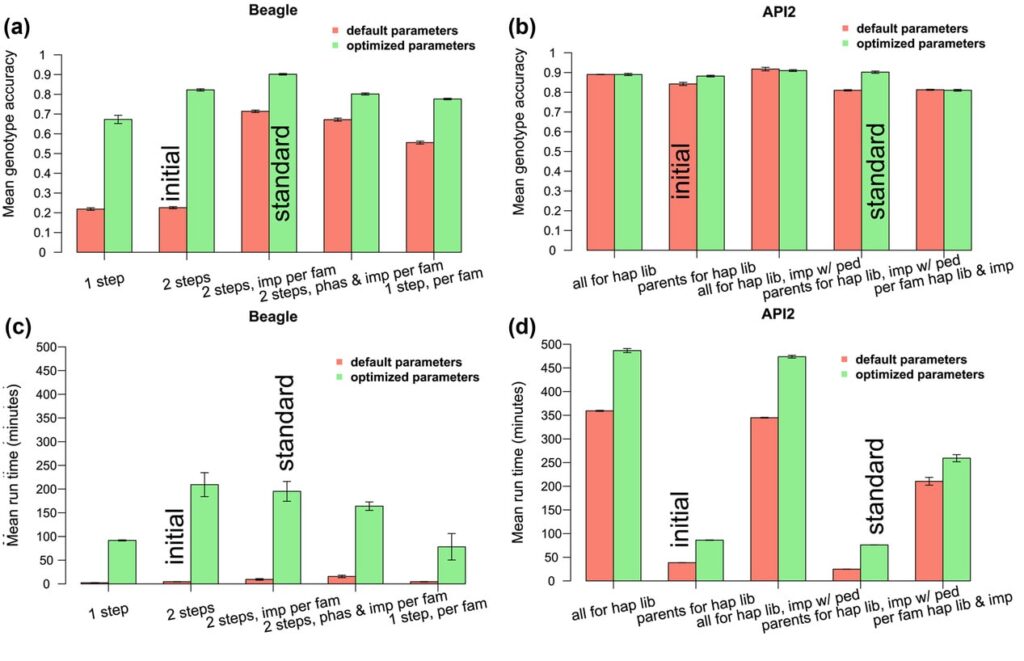
Beagle is a program package for Windows which allows you to phase genotypes and impute ungenotyped markers. The latest iteration showcases enhance memory utilization and computational efficiency, particularly beneficial for the analysis of extensive sequence datasets.
General notes
To run the application, you need to have Java version 1.8 or later installed. You can verify the deployment by entering a special command prompt. The latest interpreter can be downloaded from the official website. Please note that attempting to launch Beagle with an outdated version will result in an error.
The utility relies on a customized iteration of the Li and Stephens haplotype frequency model. It optimizes both computational efficiency and storage requirements. The algorithm is documented in various scientific papers and has gained widespread adoption in the field of population genetics.
Input and output
The software employs Variant Call Format for both input and output of genotype data. To ensure accurate analysis, it is essential that pseudoautosomal and non pseudoautosomal X chromosome genotypes are segregated into distinct input files and processed separately.
Two output items are generated. The log file provides a comprehensive summary of the analysis. It encompasses details such as the program version, command line arguments, and the computational time involved. To perform haplotype analysis, you may try a tool called Haploview.
Features
- free to download and use;
- compatible with modern Windows versions;
- enables you to phase and impute genotypes;
- you can enter various command line arguments;
- it is possible to specify multiple parameters.