With the help of this specialized application biologists can visualize and analyze custom phylogenetic trees in different formats like Newick or Nexus.
FigTree
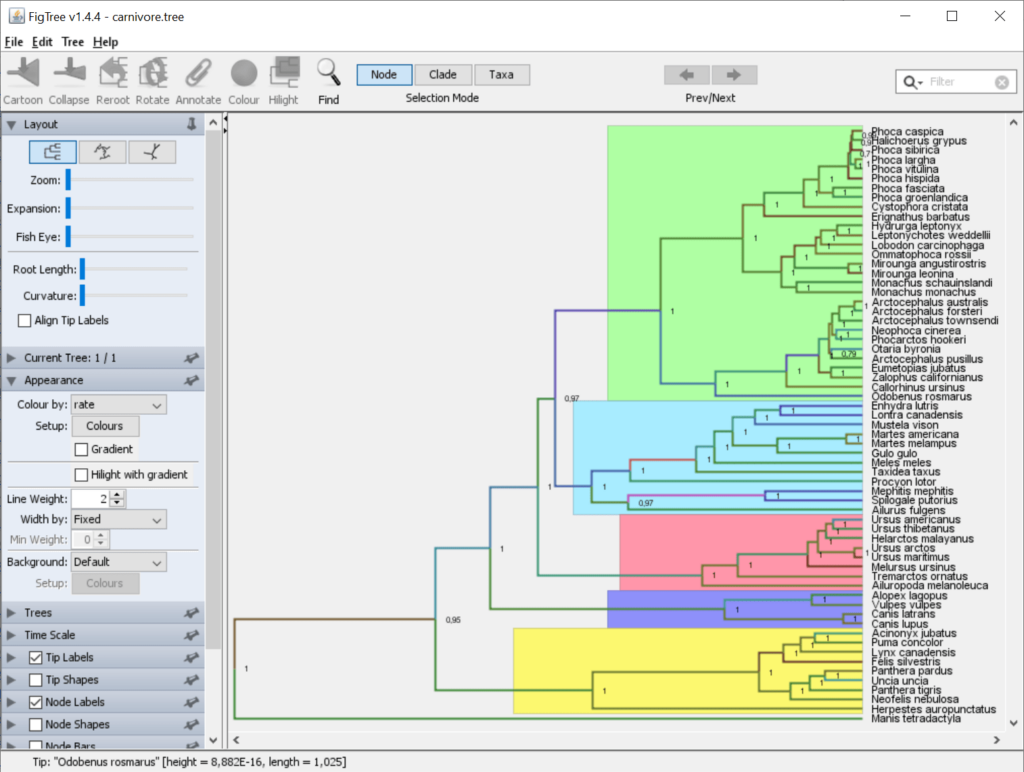
FigTree is a Windows software oriented toward professional scientific researchers in the fields of biology and bioinformatics. It empowers users to explore phylogenetic datasets and present resulting hierarchical structures in a visually appealing manner. Like in PAUP, there is an option to display node labels and branch lengths to suit specific needs.
Tree analysis
After launching the utility you can open an existing project through the File menu. It is possible to hide or display various visual elements such as:
- Tip labels;
- Node shapes;
- Scale bar;
- Legend, etc.
There are sliders for zooming in and out of the tree as well as for tweaking the root length. Moreover, users are able to change the line curvature and activate the fish eye effect. This is especially advantageous when working with extensive datasets.
Editing and annotation
Instruments to select the preferred color scheme and line weight are provided. Additionally, you can insert personal notes into the tree. Finished projects may be exported to several formats, including PDF, SVG, PNG and JPEG, which is crucial for effortless sharing with other people.
Features
- free to download and use;
- contains tools for interacting with complex phylogenetic trees on the desktop;
- you can add new nodes and annotate existing structures;
- there are flexible background color and transparency settings;
- supports multiple output formats, simplifying further manipulations;
- compatible with all modern versions of Windows.